Here is a list of bioinformatics software written by the Hammell Lab:
Published
TEtranscripts
TEtranscripts is a software package that utilizes both unambiguously (uniquely) and ambiguously (multi-) mapped reads to perform differential enrichment analyses from RNA-seq experiments.
TEsmall
TEsmall is a tool that allows for the simultaneous processing and analysis of a variety of small RNAs in a single integrated workflow.
SAKE
SAKE is a software package that utilizes a robust method for scRNA-seq analysis, and provides quantitative statistical metrics at each step of the analysis pipeline
Beta
TEpeaks
TEpeaks is a software package for including repetitive regions in peak calling from ChIP-seq datasets.
TElocal
TElocal is a software package for quantifying transposable elements at the locus level from RNA-seq datasets.
ezBAMQC
ezBAMQC is a software package that performs quality control on alignment files from high throughput sequencing experiments and assesses their suitability for downstream analysis.
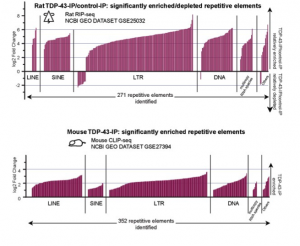 TDP-43 is an RNA-binding protein that is known to control proper splicing and translation of many RNA targets in neurons. Mutation of TDP-43 has been associated with a variety of neurdogenerative diseases including Amyotrophic Lateral Sclerosis (ALS), Fronto-Temporal Lobar Degeneration (FTLD), and Alzheimer’s Disease (AD). However, the normal function of TDP-43 in neuronal development and maintenance has not been fully characterized and few of its mRNA targets have been definitively associated with the neurodegenerative diseases that result from loss of TDP-43 function. In collaboration with the Dubnau lab at CSHL, my group has explored the novel hypothesis that TDP-43 normally plays a large and hitherto uncharacterized role in regulating the expression of transposable elements (TEs), mobile genetic elements whose unregulated expression leads to genetic instability as well as cellular toxicity. Members of my group have shown that TDP-43 binds widely to TE transcripts in mammals, and that TDP-43 binding to TEs is lost in human patients diagnosed with FTLD (Li et al., 2012), a disease characterized by TDP-43 proteinopathy. While these studies support a role for TDP-43 in regulating TE expression, our future goals are centered on defining a causal role for TDP-43 mediated regulation of TEs in neurodegenerative disease. One important element of this project will be the identification of how TDP-43 interacts with the small RNA regulators of TE expression known to be involved in controlling TE mobility.
TDP-43 is an RNA-binding protein that is known to control proper splicing and translation of many RNA targets in neurons. Mutation of TDP-43 has been associated with a variety of neurdogenerative diseases including Amyotrophic Lateral Sclerosis (ALS), Fronto-Temporal Lobar Degeneration (FTLD), and Alzheimer’s Disease (AD). However, the normal function of TDP-43 in neuronal development and maintenance has not been fully characterized and few of its mRNA targets have been definitively associated with the neurodegenerative diseases that result from loss of TDP-43 function. In collaboration with the Dubnau lab at CSHL, my group has explored the novel hypothesis that TDP-43 normally plays a large and hitherto uncharacterized role in regulating the expression of transposable elements (TEs), mobile genetic elements whose unregulated expression leads to genetic instability as well as cellular toxicity. Members of my group have shown that TDP-43 binds widely to TE transcripts in mammals, and that TDP-43 binding to TEs is lost in human patients diagnosed with FTLD (Li et al., 2012), a disease characterized by TDP-43 proteinopathy. While these studies support a role for TDP-43 in regulating TE expression, our future goals are centered on defining a causal role for TDP-43 mediated regulation of TEs in neurodegenerative disease. One important element of this project will be the identification of how TDP-43 interacts with the small RNA regulators of TE expression known to be involved in controlling TE mobility.
 Amyotrophic Lateral Sclerosis (ALS) is a fatal progressive neurodegenerative disorder with no known cure, and only two FDA-approved treatments that appear to mildly slow disease progression. ALS is a largely sporadic disease, with 90% of patients carrying no known genetic mutation or family history of disease. Large-scale patient sequencing studies have identified a growing number of genes in which mutations are linked to ALS, with the most common being a repeat expansion in the intronic region of C9orf72.
Amyotrophic Lateral Sclerosis (ALS) is a fatal progressive neurodegenerative disorder with no known cure, and only two FDA-approved treatments that appear to mildly slow disease progression. ALS is a largely sporadic disease, with 90% of patients carrying no known genetic mutation or family history of disease. Large-scale patient sequencing studies have identified a growing number of genes in which mutations are linked to ALS, with the most common being a repeat expansion in the intronic region of C9orf72.
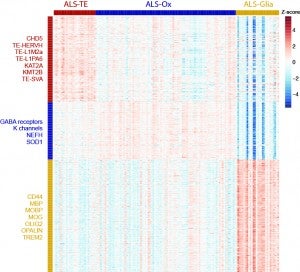
In collaboration with Target ALS and the New York Genome Center ALS Consortium, we are analyzing the transcriptomic profiles of post-mortem brain tissues from ALS patients to determine if there are potential subtypes in patient samples. Using TEToolkit and SAKE, we were able to identify three distinct clusters of gene expression profiles in ALS frontal and motor cortex samples. The largest cluster, identified in 61% of patient samples, displayed hallmarks of oxidative and proteotoxic stress. Another 20% of the ALS patient samples exhibited high levels of retrotransposon expression and other signatures of TDP-43 dysfunction. Finally, a third group showed predominant signatures of glial activation (19%). We aim to correlate these molecular subtypes with clinical presentation and disease progression, while also determining if these (or other) subtypes exist in other tissues, such as spinal cord and muscle. The project would also expand to other neurodegenerative diseases to determine if transposable element upregulation could be an informative indicator for these disorders.
