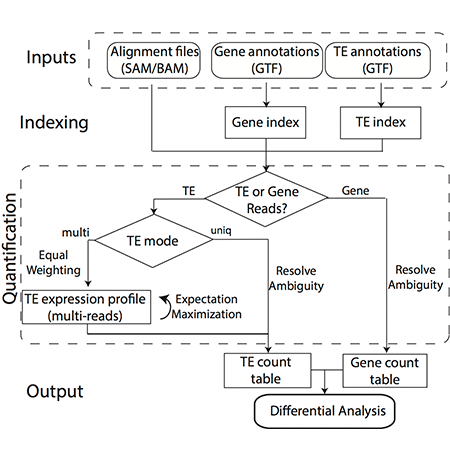
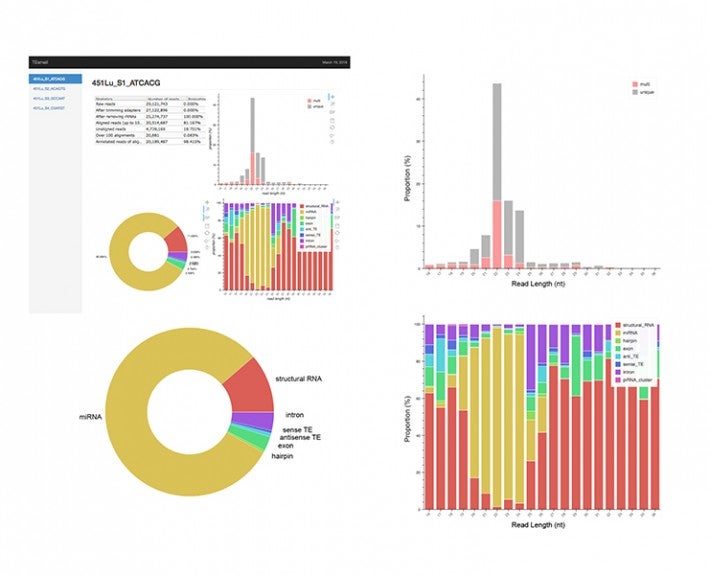
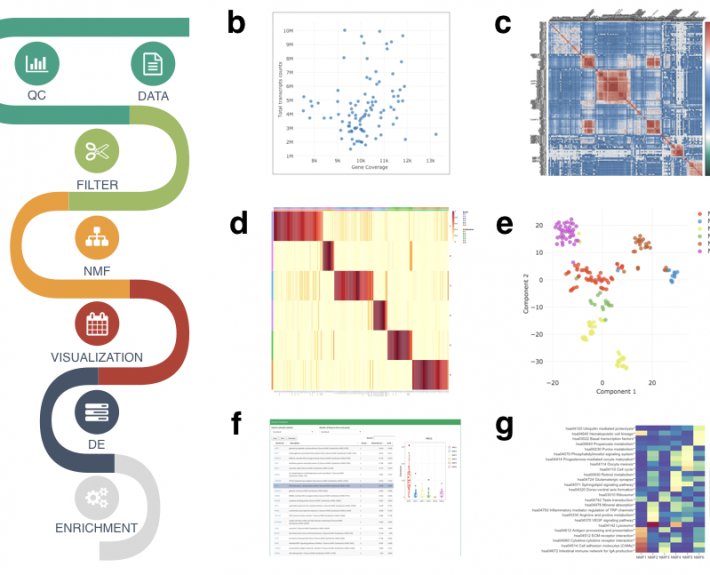
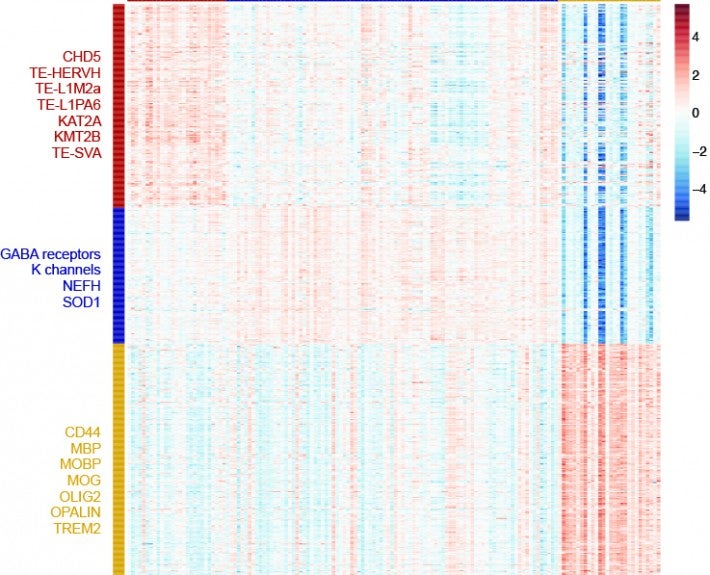
To ensure that cells function normally, tens of thousands of genes must be turned on or off together. To do this, regulatory molecules – transcription factors, RNA binding proteins and non-coding RNAs – simultaneously control hundreds of genes. The Gale-Hammell lab studies how the resulting gene networks function and how they can be compromised in human disease. This includes an emphasis on developing novel tools for the statistical analysis of genomics data, developing novel algorithms for modeling the flow of signals through genetic pathways, and importantly, testing these models using the tools of molecular genetics.
Molly Gale Hammell
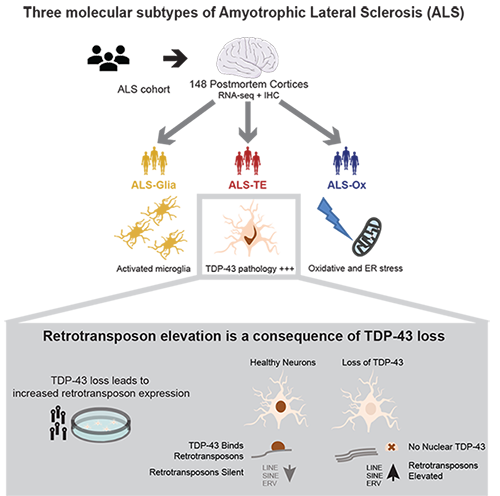
Transposable Elements; Amyotrophic Lateral Sclerosis (ALS); RNA biology; Small RNAs; Computational Genomics; Neurodegenerative Disease
External funding sources
We wish to acknowledge the following groups for providing us additional funding for our research.